Rapid and efficient COVID-19 testing has been a pressing concern since the onset of the pandemic. Various technologies have been employed in the detection of viral RNA or proteins, or the antibodies generated against them mucous or other easily collected bodily fluids. CRISPR-Cas technology has been similarly exploited to detect RNA extracted from nasopharyngeal swabs. The viral genes are first amplified by RT-LAMP before Cas12a binds with the complimentary ssRNA, leading to collateral cleavage of ssDNA fluorescent reporter molecules. This method takes only 40 minutes, compared with the several hours required by RT-qPCR, and necessitates less sophisticated equipment.
Nasopharyngeal swabs ensure a high-quality collection of viral RNA, though they are considered unpleasant for the individual being tested and potentially risky for the collector, requiring close contact. Further, processing and extracting the RNA from these samples then requires costly reagents, usually from expensive commercially sourced kits. Some studies have suggested that these samples could, instead, be directly lysed and then used in molecular diagnostic assays without the use of these kits, hugely reducing the cost of analysis for low and middle-income countries in particular.
In a paper recently uploaded to the preprint server medRxiv* by Edward Málaga Trillo et al. (April 29th, 2021), these methods are tested in combination with CRISPR-Cas detection in the field, demonstrating a sensitivity and specificity of 93.8% and 99%, respectively.
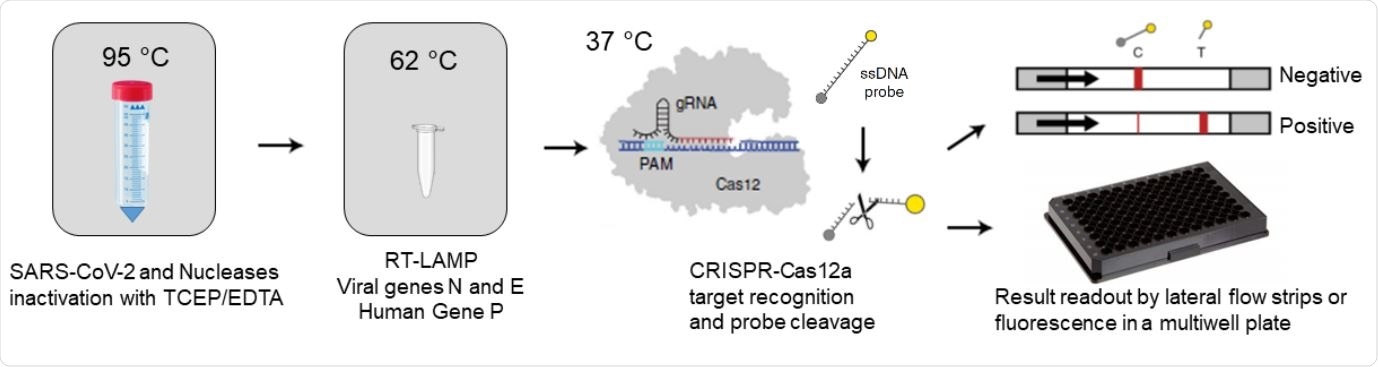
RCSMS detection workflow. Upon saliva treatment with TCEP/EDTA, 2 µl of inactivated sample are added to a 10 µl RT-LAMP reaction. A 2 µl aliquot of the RT-LAMP product is then mixed with the RNP complex consisting of Cas12 and RNA guides. Recognition of viral target sequences by the RNP complex triggers the collateral activity of Cas12a, resulting in the cleavage of ssDNA reporter probes. For immunocromatographyic (qualitative) readout, a lateral flow strip is then inserted into the CRISPR-Cas12a reaction tube or well. Within two minutes, uncleaved reporter molecules flow and accumulate into the control capture line of the strip (C band in the image), whereas cleaved reporter molecules flow towards the target capture line of the strip (T band in the image), (adapted from Broughton et al. (8) and Patchsung et al. (18)). For fluorescence (quantitavite) readout, CRISPR-Cas12a reactions are recorded in real-time over 10 min using an automated plater reader; cleaved reporter molecules yield a bright fluorescent signal
How was the method tested?
Saliva and nasopharyngeal swab samples were collected from 276 individuals that had reported severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection-related symptoms, with those that had received a vaccine or failing other particular criteria being excluded. Any viral RNA in the samples was inactivated using a solution of tris(2-carboxyethyl)phosphine hydrochloride (TCEP-HCl), EDTA, and sodium hydroxide, which subsequently go through a single heating-cooling cycle, and then the denatured RNA was extracted. RT-LAMP amplification was then performed for 30 minutes at 62⁰C before the CRISPR-Cas system was applied.
Viral genes N (934 bp) and E (540 bp), and the human gene POP7 (406 bp) were selected for RT-LAMP amplification, and the complimentary guide RNA-Cas12a enzyme to these genes were prepared. The reactions were allowed to run for ten minutes in the presence of 50 nM of the Cas12a-guide RNA complex.
Of the 276 samples, 28.3% were positive for the E gene while 29.3% were positive for the N gene, though two false positives and five false negatives were later detected. The limit of detection of the system was tested by intentional doping with synthetic RNA, finding that the RNA was detectable at only 5 viral copies per 10 µL of sample, 2 µL being the inactivation solution.
The method demonstrated a good level of agreement with RT-qPCR tests, both in the laboratory and in the field, and saliva samples were also consistent with CRISPR-Cas detection systems based on nasopharyngeal swaps alone. As lysis and inactivation of the virus occur immediately following collection, there is a much lower risk of transmission, and biosafety level 2 laboratories are not required for testing. The system can be easily reprogrammed in the event of mutations to the virus genome that makes the currently selected guide RNAs ineffective. Other similar CRISPR-Cas systems have previously been developed that target the SARS-CoV-2 spike protein, utilizing Cas13a. The group also attempted the same detection method against a section of the spike protein with limited success, suggesting that the Cas12a enzyme utilized is unsuitable. It usually targets transcribed post-amplification RNA.
This study was performed in the context of widespread COVID-19 in Peru combined with a limited testing capacity and aims to provide a cost-effective method of clinical validation. The test methodology developed by the group termed rapid coronavirus sensitive monitoring from saliva (RCSMS), can easily be applied at both the primary care level and in diagnostic laboratories and could be an important tool in detecting SARS-CoV-2 in less developed countries in particular.
*Important Notice
medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Journal reference:
- Clinical validation of RCSMS: a rapid and sensitive CRISPR-Cas12a test for the molecular detection of SARS-CoV-2 from saliva, Joaquín Abugattás Núñez del Prado, Angélica Quintana Reyes, Juan Blume La Torre, Renzo Gutiérrez Loli, Alejandro Pinzón Olejua, Elena Rocío Chamorro Chirinos, Félix Antonio Loza Mauricio, Jorge L. Maguiña, Julio Leon, Piere Rodríguez Aliaga, Edward Málaga Trillo, medRxiv, 2021.04.26.21256081; doi: https://doi.org/10.1101/2021.04.26.21256081, https://www.medrxiv.org/content/10.1101/2021.04.26.21256081v2









 Add Category
Add Category

